Principle Components Analysis (PCA)
TL;DR
The usual procedure to calculate the -dimensional principal component analysis consists of the following steps:
Calculate
average
data matrix
scatter matrix (covariance matrix)
of all feature vectors
Calculate the normalized () eigenvectors and sort them such that the corresponding eigenvalues are decreasing, i.e.
Construct a matrix
with the first eigenvectors as its columns
Transform each feature vector into a new feature vector
of smaller dimension
Dimensionality reduction
Goal: represent instances with fewer variables
- Try to preserve as much structure in the data as possible
- Discriminative: only structure that affects class separability
Feature selection
- Pick a subset of the original dimensions
- Discriminative: pick good class “predictors”
Feature extraction
Construct a new set of dimensions
- : features
(Linear) combinations of original
Direction of greatest variance
- Define a set of principal components
- 1st: direction of the greatest variability in the data (i.e. Data points are spread out as far as possible)
- 2nd: perpendicular to 1st, greatest variability of what’s left
- …and so on until (original dimensionality)
- First components become dimensions
- Change coordinates of every data point to these dimensions

Q: Why greatest variablility?
A: If you pick the dimension with the highest variance, that will preserve the distances as much as possible
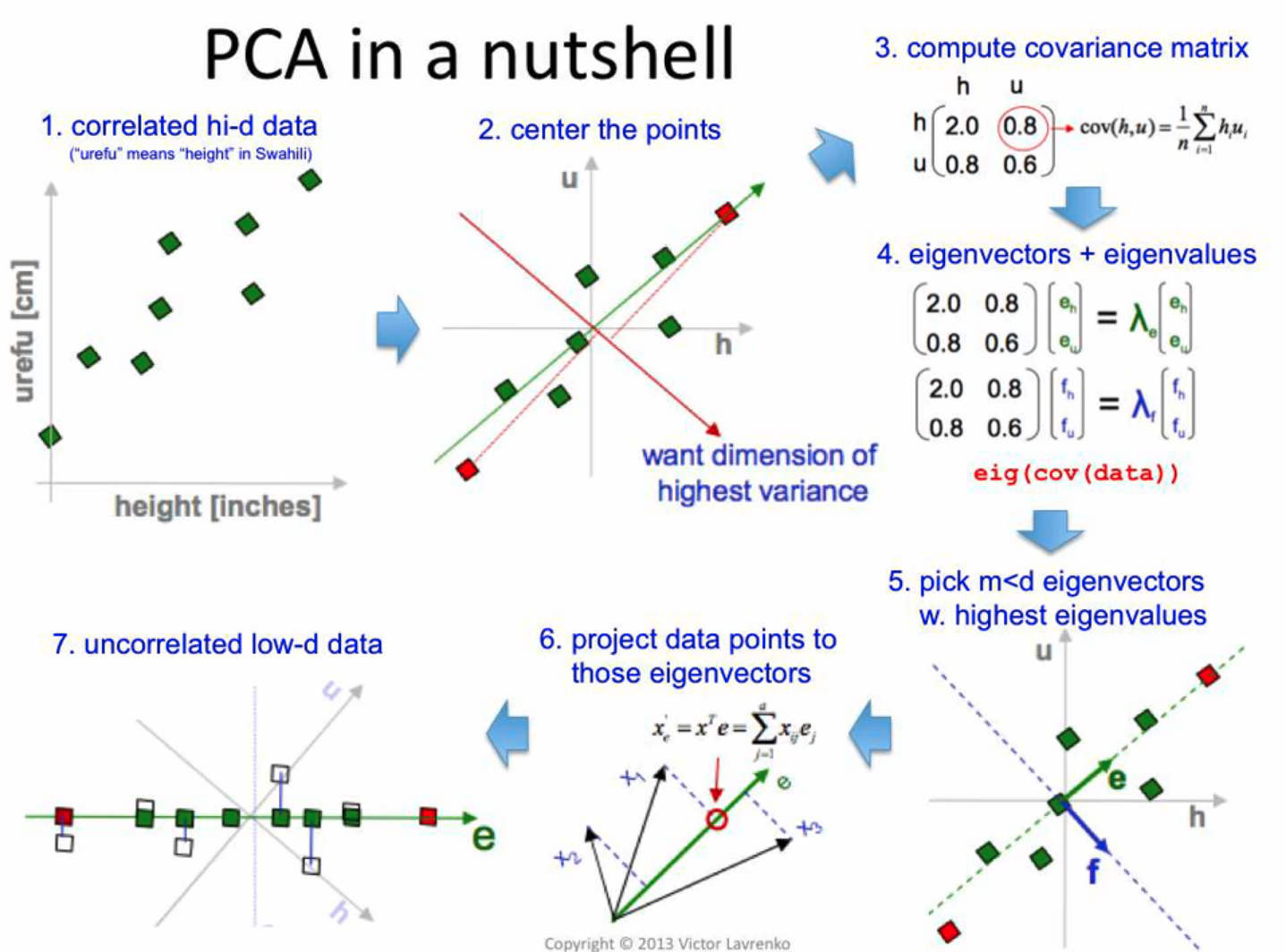
How to PCA?
“Center” the data at zero (subtract mean from each attribute)
Compute covariance matrix
The covariance between two attributes is an indication of whether they change together (positive correlation) or in opposite directions (negative correlation).
For example, When increases/decreases, also increases/decreases.
We want vectors which aren’t turned by covariance matrix :
are eigenvectors of , and are corresponding eigenvalues
Principle components = eigenvectors with largest eigenvalues
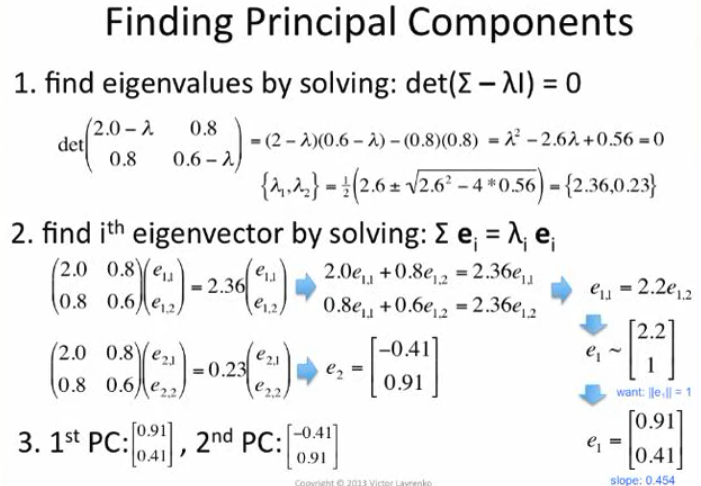
Finding principle components
Find eigenvalues by solving Characteristic Polynomial
- : Identity matrix
Find -th eigenvector by solving
and we want to have unit length ()
Eigenvector with the largest eigenvalue will be the first principle component, eigenvector with the second largest eigenvalue will be the second priciple component, so on and so on.
Example
Projecting to new dimension
We pick $m
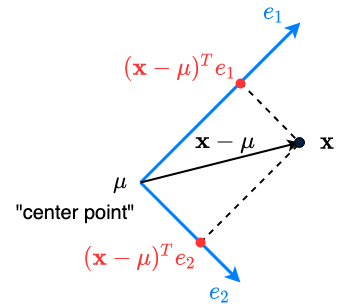
For instance (original coordinates), we want new coordinates
- “Center” the instance (subtract the mean):
- “Project” to each dimension: for
Example
Go deeper in details
Why eigenvectors = greatest variance?
Why eigenvalue = variance along eigenvector?
How many dimensions should we reduce to?
Now we have eigenvectors and we want new dimension
We pick that “explain” the most variance:
Sort eigenvectors s.t.
Pick first eigenvectors which explain 90% or the total variance (typical threshold values: 0.9 or 0.95)
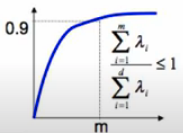
Or we can use a scree plot
PCA in a nutshell
PCA example: Eigenfaces
Perform PCA on bitmap images of human faces:
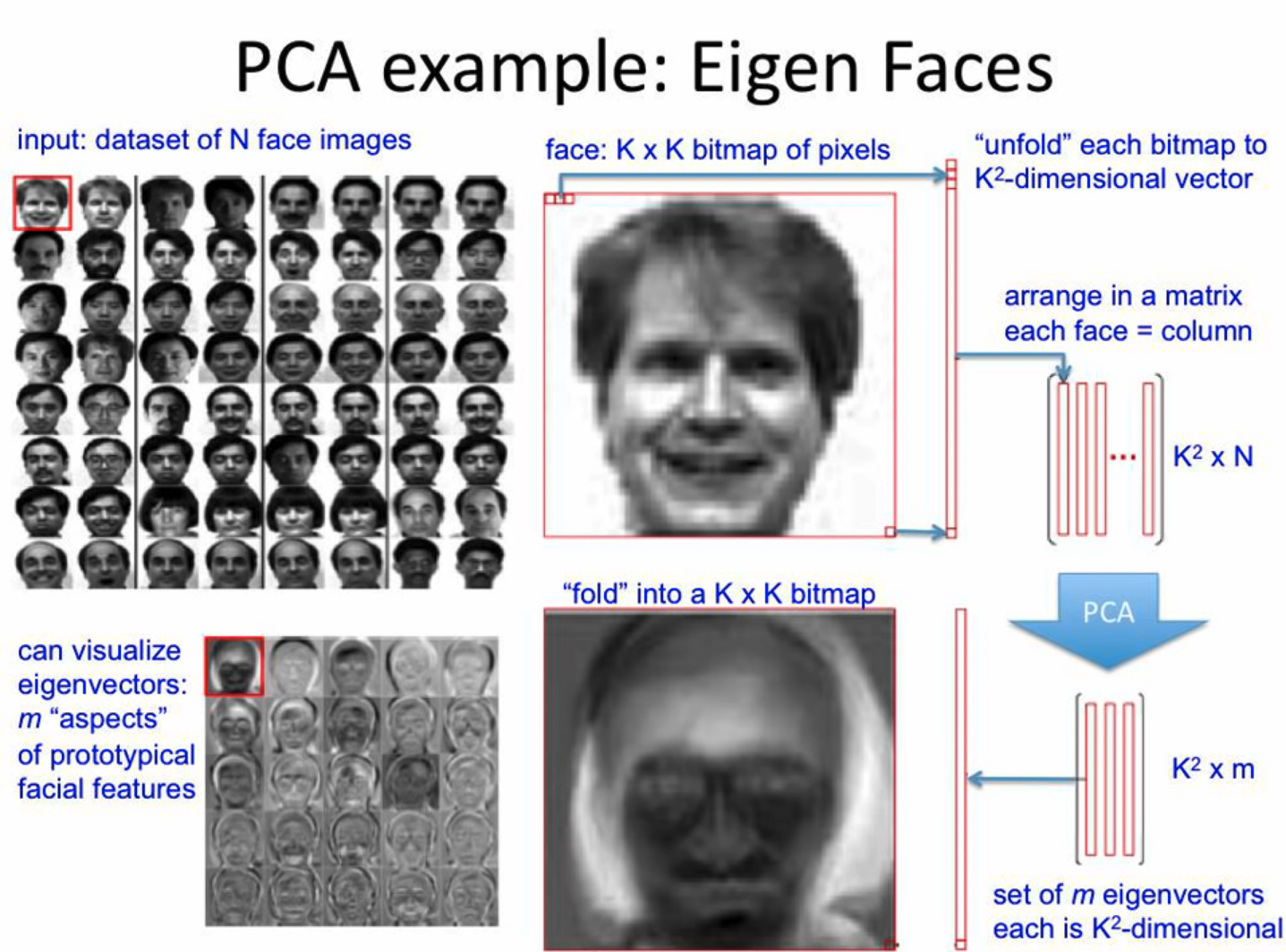
Belows are the eigenvectors after we perform PCA on the dataset:

Then we can project new face to space of eigen-faces, and represent vector of new face as a linear combination of principle components.

As we use more and more eigenvectors in this decomposition, we can end up with a face that looks more and more like the original guy

Why is eigenface neat and interesting?
- This is neat because by taking the first few eigenvectors you can get a pretty close representation of the face. Suppose that this corresponds to maybe 20 eigenvectors. This means you’re using only 20 numbers to represent a face bitmap which looks kind of like the original guy! Can you use only 20 pixels to represent him nearly? No, there’s no way!
- You’re effectively picking 20 numbers/mixture coefficients/coordinates. One really nice way to use this is you can use this for massive compression of the data. If you communicate to others if they all have access to the same eigenvectors, all they need to send between each other are just the projection coordinates. Then they can transmit arbitrary faces between them. This is massive reduction in the size of data.
- Your classifier or your regression system now operate in low dimensional space. So they have plenty of redundancy to grab on to and learn a better hyperplane. 👏
Application of eigenface
Face similarity
- in the reduced space
- insensitive to lighting expression, orientation
Projecting new “faces”

Pratical issues of PCA
PCA is based on covariance matrix and covariance is extremely sensitive to large values
E.g. multiple some dimension by 1000. Then this dimension dominates covariance and become a principle component.
Solution: normalize each dimension to zero mean and unit variacne
PCA assumes underlying subspace is linear.
PCA can sometimes hurt the performace of classification
Because PCA doesn’t see the labels
Solution: Linear Discriminant Analysis (LDA)
Picks a new dimension that gives
- maximum separation between means of prejected classes
- minimum variance within each projected class
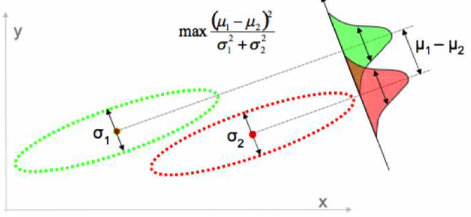
But this relies on some assumptions of the data and does not always work. 🤪
Reference
- Principle Component Analysis: a great series of video tutorials explaining PCA clearly 👍