Numpy Random
Common used functions
import numpy as np
We use numpy.random package to generate an array of random values instead of looping through the random generation of one variable.
np.random.randn()
Return a sample (or samples) from the “standard normal” distribution.
Example
np.random.randn(2,4)[[-0.50516164 0.03107622 -1.98470915 -0.06278207] [ 0.00806484 1.60814316 -0.06865081 0.90962803]]
np.random.rand()
Create an array of the given shape and populate it with random samples from a uniform distribution over
[0, 1).Example
np.random.rand(2,4)[[0.93885544 0.0643788 0.74463388 0.97446713] [0.03621414 0.0420926 0.54597933 0.72757245]]
np.random.randint()
Return random integers from the “discrete uniform” distribution in the “half-open” interval [low, high). If high is None (the default), then results are from [0, low).
Example
np.random.randint(0, 10, size=(2, 3))array([[2, 6, 0], [1, 5, 8]])
np.random.permutation()
Randomly permute a sequence, or return a permuted range.
Example
np.random.permutation([1, -4, 3, 2, -6])[-6 2 1 3 -4]
np.random.seed()
The random algorithm used for all of the methods above is a pseudo random generating algorithm. It is based on some initial state, or “seed” to generate random numbers.
- If you do not specify the seed, it can take some number elsewhere to be the seed.
- But if we specify the same seed every time we generate random numbers, those numbers will be the same. It is good for many cases, for example, replicating exactly the result of your neural network training even you randomly initialized your weights.
Example:
print("When we do not specify the seed:")
for i in range(3):
print(np.random.randint(0,10))
print("When we specify the same seed:")
for i in range(3):
np.random.seed(1111) # set the same seed 1111 before every random generation
print(np.random.randint(0,10))
print("When we specify different seeds:")
for i in range(3):
np.random.seed(i * 3)
print(np.random.randint(0,10))
When we do not specify the seed:
9
5
5
When we specify the same seed:
7
7
7
When we specify different seeds:
5
8
9
Random initialization
For a neural network to learn well, beside feature normalization and other things, we also need proper weight initialization:
- the weights should be randomly initialized,
- or at least different numbers (to break the symmetry),
- and they should be small.
There’re two good methods for good initialization. They both take the size of the layers into account (and to be more precisely, they also consider the activation functions).
Xavier initialization
It’s good when use with sigmoid or tanh activation functions.
- : the number of neurons from the previous layer
- : the number of neurons from the current layer
n_in = 3 # the number of neurons from the previous layer
n_out = 2 # the number of neurons from the current layer
alpha = np.sqrt(2. / (n_in + n_out))
W = np.random.randn(n_out, n_in) * alpha
print(W)
[[-0.64820888 1.28386704 0.08342666]
[ 1.0910627 1.16949752 0.63448025]]
Summary:
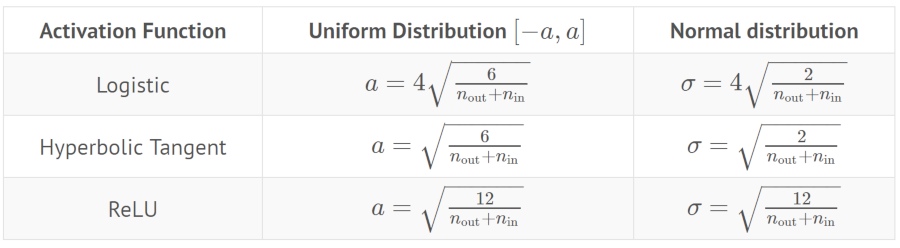
He initialization
It’s good when use with ReLU activation function.
Sampling
np.random.choice()
Generates a random sample from a given 1-D array
Example
def sm_sample_general(out, smp): # out contains possible outputs # smp contains the softmax output distributions return np.random.choice(out, p = smp) out = ['a', 'b', 'c'] smp=np.array([0.3, 0.6, 0.1]) outputs = [] for i in range(10): outputs.append(sm_sample_general(out, smp)) print(outputs) outputs = [] # Law of large numbers: 100000 is large enough for our sample to approximate the true distribution for i in range(100000): outputs.append(sm_sample_general(out, smp)) from collections import Counter c_list = Counter(outputs) print(c_list)['c', 'b', 'b', 'a', 'c', 'a', 'b', 'b', 'a', 'b'] Counter({'b': 60044, 'a': 29928, 'c': 10028})
Dropout
Act as a regularization, aimming to make the network less prone to overfitting.
In training phase, with Dropout, at each hidden layer, with probability
p, we kill the neuron.What it means by ‘kill’ is to set the neuron to 0. As neural net is a collection multiplicative operations, then those 0 neuron won’t propagate anything to the rest of the network.

Let
nbe the number of neuron in a hidden layer, then the expectation of the number of neuron to be active at each Dropout isp*n, as we sample the neurons uniformly with probabilityp.- Concretely, if we have 1024 neurons in hidden layer, if we set
p = 0.5, then we can expect that only half of the neurons (512) would be active at each given time. - Because we force the network to train with only random
p*nof neurons, then intuitively, we force it to learn the data with different kind of neurons subset. The only way the network could perform the best is to adapt to that constraint, and learn the more general representation of the data.
- Concretely, if we have 1024 neurons in hidden layer, if we set
Implmentation
Sample an array of independent Bernoulli Distribution, which is just a collection of zero or one to indicate whether we kill the neuron or not.
- If we multiply our hidden layer with this array, what we get is the originial value of the neuron if the array element is 1, and 0 if the array element is also 0.
Scale the layer output with
pNecause we’re only using
p*nof the neurons, the output then has the expectation ofp*x, ifxis the expected output if we use all the neurons (without Dropout).As we don’t use Dropout in test time, then the expected output of the layer is
x. That doesn’t match with the training phase. What we need to do is to make it matches the training phase expectation, so we need to scale the output withp
# Dropout training, notice the scaling of 1/p u1 = np.random.binomial(1, p, size=h1.shape) / p h1 *= u1